Short Description
Jep embeds CPython in Java through JNI.
Code on Python through Java !
Institution: Institut Pasteur
Website: https://research.pasteur.fr/en/team/bioimage-analysis/
Documentation
Some benefits of embedding CPython in a JVM:
- Using the native Python interpreter may be much faster than alternatives.
- Python is mature, well-supported, and well documented.
- Access to high quality Python modules, both native CPython extensions and Python-based.
- Compilers and assorted Python tools are as mature as the language.
- Python is an interpreted language, enabling scripting of established Java code without requiring recompilation.
- Both Java and Python are cross-platform, enabling deployment to different operating systems.
Installation instructions
Simply run pip install jep or download the source and run python setup.py build install. Building and installing require the JDK, Python, and optionally numpy to be installed beforehand.
On your Icy plugin project, add this dependency
<dependency>
<groupId>org.bioimageanalysis.icy</groupId>
<artifactId>icy-jep</artifactId>
<version>4.1.0</version>
</dependency>
Dependencies
- Python >= 3.5
- Java >= 1.8
- NumPy >= 1.7 (optional)
Notable features
- Interactive Jep console much like Python’s interactive console
- Supports multiple, simultaneous, mostly sandboxes sub-interpreters or shared interpreters
- Numpy support for Java primitive arrays
Help
We welcome comments, contributions, bug reports, wiki documentation, etc.
If you need help, please first search for existing solutions online, in the issues, and on the wiki. If you still need help, please open a GitHub issue, and we will try and help you. Please remember to close the issue once it has been resolved.
Jep Team
Citation
Please also cite the Icy software and mention the version of Icy you used (bottom right corner of the GUI or first lines of the Output tab):
de Chaumont, F. et al. (2012) Icy: an open bioimage informatics platform for extended reproducible research, Nature Methods, 9, pp. 690-696
https://icy.bioimageanalysis.org
Author(s)
JEP Team Nate Jensen & Ben Steffensmeier
Plugin conversion
Amandine Tournay & Carlos Garcia Lopez de Haro
Usage
To enable this plugin in Icy, you need to configure the path to your selected Python.
Go in the search bar and search for Python3Preferences.
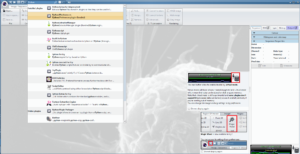
If you are a user of Conda, you select one of the environments available:
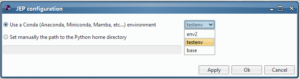
Or if you are a user of a classical Python installation or if you are using a Virtual environment, you can select the home directory
of your Python:
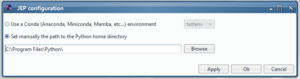
There you are !
Click on apply or save, and you are ready to use Python in Icy !
Utility functions
Reminder: When you open a new Python instance, do NOT FORGET to close when you are done ! 😀
try {
// Open a Python instance
SubInterpreter python = new PythonExec().getInterpreter();
// Same but adding a JEP configuration object to manage better the link between Java and Python (see JavaDoc)
SubInterpreter python2 = new PythonExec(new JepConfig()).getInterpreter();
// Running some Python code
python2.exec("x = 5");
python2.runScript("<Python script file path>");
// Prints 5
System.out.println(python2.getValue("x"));
// Send data to Python
python2.set("y", 10);
// Prints 10 on the Python output stream
python2.exec("print(y)");
// Close a Python instance
python.close();
// Always close an opened instance !
python2.close();
}
catch (JepException e) {
IcyExceptionHandler.showErrorMessage(new JepException(e), true, true);
}