Short Description
This plugin ships automated methods for extracting trajectories of multiples objects in a sequence of 2D or 3D images. Up to version 2 it was known as the ‘Probabilistic particle tracker’ plugin.
A number of precious features for object tracking in microscopy images are embedded: the number of targets can vary through time (objects can appear and disappear), false detection (not originating from a target) are automatically detected and discarded, different target dynamics are available (diffusive, directed movement, or both).
The tracking methods relies on a published method (Chenouard et al., TPAMI, 2013) which is termed ‘multiframe’ in the sense that at a given time point of the image sequence multiple past and futures frames are considered for building the best set of tracks. It is part of the family of ‘probabilistic’ tracking methods in the sense that a statistical model of the particle trajectories and acquisition device is built such as tracking decisions are optimal according to a given statistical criterion with respect to this model. By default, a rough estimation of the parameters of the statistical model will be evaluated by the software such that virtually no parameter tuning is required from the user. However, for optimal control of the methods, a dedicated interface can give access to the full set of parameters.
The tracking methods are freely provided for use and modification. By using the provided methods you agree to properly reference the scientific work at their roots in any written or oral communication exposing a work that took advantage of them. The exact reference is: Nicolas Chenouard, Isabelle Bloch, Jean-Christophe Olivo-Marin, “Multiple Hypothesis Tracking for Cluttered Biological Image Sequences,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 11, pp. 2736-3750, Nov., 2013
Pubmed link: http://www.ncbi.nlm.nih.gov/pubmed/23689865
Documentation
Summary
This plugin ships automated methods for extracting trajectories of multiples objects in a sequence of 2D or 3D images.
A number of precious features for object tracking in microscopy images are embedded: the number of targets can vary through time (objects can appear and disappear), false detection (not originating from a target) are automatically detected and discarded, different target dynamics are available (diffusive, directed movement, or both).
The tracking methods relies on a published method (Multiple Hypothesis Tracking algorithm, aka MHT, described in Chenouard et al., TPAMI, 2013) which is termed ‘multiframe’ in the sense that at a given time point of the image sequence multiple past and futures frames are considered for building the best set of tracks. It is part of the family of ‘probabilistic’ tracking methods in the sense that a statistical model of the particle trajectories and acquisition device is built such as tracking decisions are optimal according to a given statistical criterion with respect to this model. By default, a rough estimation of the parameters of the statistical model will be evaluated by the software such that virtually no parameter tuning is required from the user. However, for optimal control of the methods, a dedicated interface can give access to the full set of parameters.
The tracking methods are freely provided for use and modification. By using the provided methods you agree to properly reference the scientific work at their roots in any written or oral communication exposing a work that took advantage of them. The exact reference is:
Nicolas Chenouard, Isabelle Bloch, Jean-Christophe Olivo-Marin, “Multiple Hypothesis Tracking for Cluttered Biological Image Sequences,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 11, pp. 2736-3750, Nov., 2013
Pubmed link: https://pubmed.ncbi.nlm.nih.gov/24051732/
Important Note:
The tracking algorithm that was used up to version 2.0 of the plugin is still accessible from the interface of the plugin by selecting Interface/Legacy Plugin in the menu of the plugin.
Manual Content
- Hands-On Manual
- Detailed description
- Developers note
- Reference
I. Hands-On Manual
In general, particle tracking analysis consists in 3 steps:
- Identification of particle locations in a time-lapse sequence. We call this step particle detection.
- Grouping detected positions through time to estimate spatio-temporal trajectories. This is the so-called particle tracking step.
- In the track analysis step, mathematical and statistical tools are used to extract relevant information from the tracks.
The Spot Tracking plugin is focused on the second step, the particle tracking task, but gives easy-access to features of ICY that allows one to perform the whole particle tracking analysis in a both efficient and accurate manner. By default, the plugin interface consists in a simple series of sections, from top to bottom, each of which allows the user to specify relevant parameters and to seamlessly make the link with particle detection and track analysis tools.
1. Particle detection
The user is first asked to specify a set of detections for the objects to track through time. To do so, detection results have to be present in the ‘swimming pool’ of ICY (an odd term designating a shared memory space between plugins). If none exist, please create one using a specialized plugin. The Spot Tracking plugin gives direct access to the Spot Detector plugin that can be used for this purpose. Just click the button ‘Run the Spot Detector plugin’ to launch it. When using this plugin don’t forget to check the option ‘Export to Swimming Pool’ on the ‘Output tab’ so that the Spot Tracking plugin can access its results.
Once detection results are present in the swimming pool, just select them in the box of the first section of the plugin. This will enable the second section if a proper set of detections is selected.
2. Particle tracking parameters
The tracking algorithms is using a complex statistical model to estimate the best trajectories from the set of particle detections. Specifying a number of parameters is thus required before extracting trajectories. Thankfully, the plugin ships a number of features that should make this step painless, even for non-expert users. One can either automatically (roughly) estimate the parameters of the model or load a predefined set of parameters used for a previous experiment.
For automatic parameter estimation, click the button ‘Estimate parameters’ and follow the directions. Please note that the estimation procedure gives only approximate parameter values that are probably sub-optimal, even though we observed satisfying results in many cases. If necessary, one can access the full set of parameters by switching to the advanced interface
(menu Interface/Advanced Interface).
Click ‘Load existing parameters‘ to import a set of existing parameters. This set of parameters needs to be stored in an XML file exported with the Spot Tracking plugin (via the menu ‘File/Save Parameters’).
3. Tracks Export for post analysis
Extracted trajectories will be automatically exported to the Track Manager plugin so that they can be visualized and various analyses and post-processes can be applied. Saving tracking results in XML files for later analysis is also performed through the menu of the Track Manager plugin.
In the third section of the plugin interface, one is asked to give a unique name for the results of each run of the plugin so that each set of results can be distinguished from one another in the Track Manager interface. Please specify in the text field a name for the result different from those already existing in the ‘swimming pool’.
Once a proper name is given, the fourth section will be made available.
4. Trajectory Extraction
Finally, one can proceed with the track extraction process per se by hitting the ‘Run tracking‘ button. Note that the process can be stopped at any time by pressing the button again.
Once tracking over the whole track sequence is completed the Track Manager plugin will be automatically launched in order to proceed with track analysis, visualization, and export
5. Troubleshoot
Spot Tracking plugin uses the LPSolve C library (see below), still in some rare cases the C library may crash (and so entirely crashed Icy which exit without any warning). In this case you can disable it from Plugin configuration menu and use the pure java solver (slower but safe) :
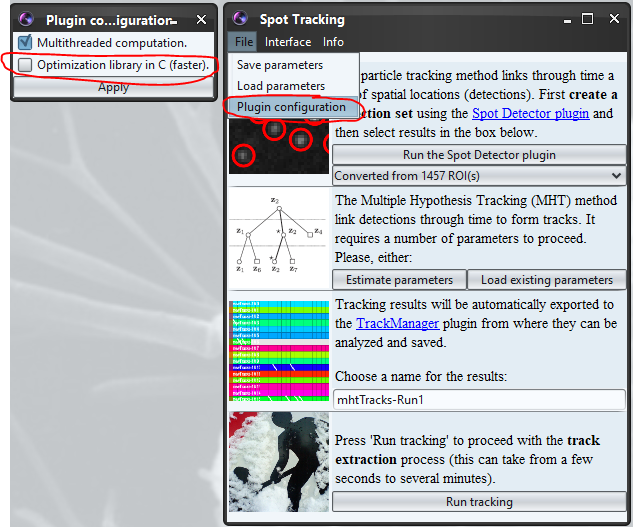
II. Detailed description
Soon to come!
Meanwhile you can refer to:
Nicolas Chenouard, Isabelle Bloch, Jean-Christophe Olivo-Marin, “Multiple Hypothesis Tracking for Cluttered Biological Image Sequences,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 11, pp. 2736-3750, Nov., 2013
for a description of the algorithm that is implemented by the plugin for track extraction. I will try soon to give a complete description here. Meanwhile, please be patient, or contact me by e-mail.
To access the whole set of parameters and options described in this publication, switch to the advanced interface using the menu: ‘Interface/Advanced Interface‘. This interface must be used to set up optimal parameters, in particular for scientific comparison between the provided algorithms and other methods since using the simplified interface yields sub-optimal results due to rough parameters estimation.
III. Developers note
The plugin is provided under licence GPLv3.0, with all sources included.
To access source files, download the plugin with the regular installer through ICY interface. Then, access the folder ‘plugins/nchenouard/particleTracking’ in the home folder of ICY. This one should contain the .jar file of the plugin. Unzip the jar file to gain access to the .java sources and .class files.
I am sorry to say that right now the documentation is pretty scarce and that a javadoc is not yet available. Please be patient, or contact by e-mail for any help with software development.
The plugin is free for any use and modifications. Please make sure that the original author and software are acknowledged if modifications are performed and that the reference [Chenouard et al., TPAMI, 2013] is cited in scientific communications making use of this work.
For the Linear Programming optization step of the MHT the default optimizer is provided by the library LPSolve 5.5 (lpsolve.sourceforge.net/5.5/). If the library cannot be loaded, a suboptimal implentation using the optimizer which can be found here: algs4.cs.princeton.edu/65reductions/Simplex.java.html is used. I am grateful to the authors of those library for publishing them.
IV. References
Nicolas Chenouard, Isabelle Bloch, Jean-Christophe Olivo-Marin, “Multiple Hypothesis Tracking for Cluttered Biological Image Sequences,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 11, pp. 2736-3750, Nov., 2013
Pubmed link: https://pubmed.ncbi.nlm.nih.gov/24051732/
6 reviews on “Spot Tracking”