Short Description
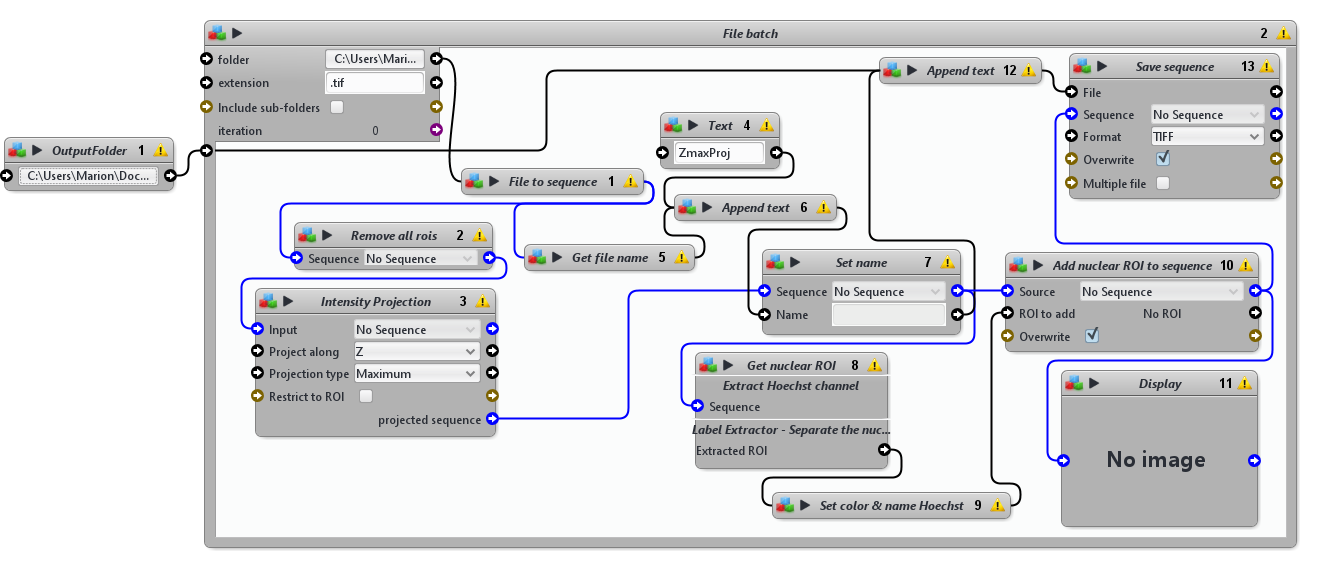
Project 3D stacks in 2D (maximum intensity projection along Z axis) and segments nuclei stained with Hoechst.
Team: Bioimage Analysis UnitInstitution: Institut Pasteur
Website:
Documentation
Please cite as: Louveaux, Marion. (2020, July 1). Nuclei segmentation from maximum intensity projections (Version 1.0). Zenodo. http://doi.org/10.5281/zenodo.3925182
Test image available at doi: 10.5281/zenodo.3925213 (choose the Jurkat cell example).
Nuclei are segmented on a maximum intensity projection of the Hoechst channel using the Icy block Gaussian filter with a radius of sigma x=5, y=5 and z=5 pixels, the Thresholder block with a manual threshold of 5000 and the Label Extractor block, which uses connected components to separate the nuclei.
This protocol is designed to loop over a folder containing 3D stacks .tif files. Input arguments to adjust in order to run this protocol: 1) in the File Batch block, provide the path to the 3D stacks .tif files folder and 2) in the OutputFolder block provide the path to the results folder, where projected and segmented images should be stored.