Short Description
Detects tissue mimics imaged with a light-sheet microscope and crops 3D sub-volume.
Related protocols: P2 – SNR and contrast measurements and P3 – Characterize the PSF from beads in 3D. Related plugin: metroloSPIM. Tissue-mimics example datasets to test the plugin and protocols are available on Zenodo.
Team: R&D Imagerie
Institution: RESTORE-GOT IT Team UMR 5070-CNRS 1301-INSERM EFS Univ. Toulouse, France
Website: https://www.itav.fr/plateformes-technologiques/R-et-D-imagerie/Team: SLN - Team Loza
Institution: ICFO-Institut de Ciences Fotonique, Av. Carl Friedrich Gauss, 3, Castelldefels, 08860, Barcelona, Spain
Website: https://www.icfo.eu/lang/research/groups/groups-details?group_id=16Citation: https://doi.org/10.1038/srep44939
Documentation
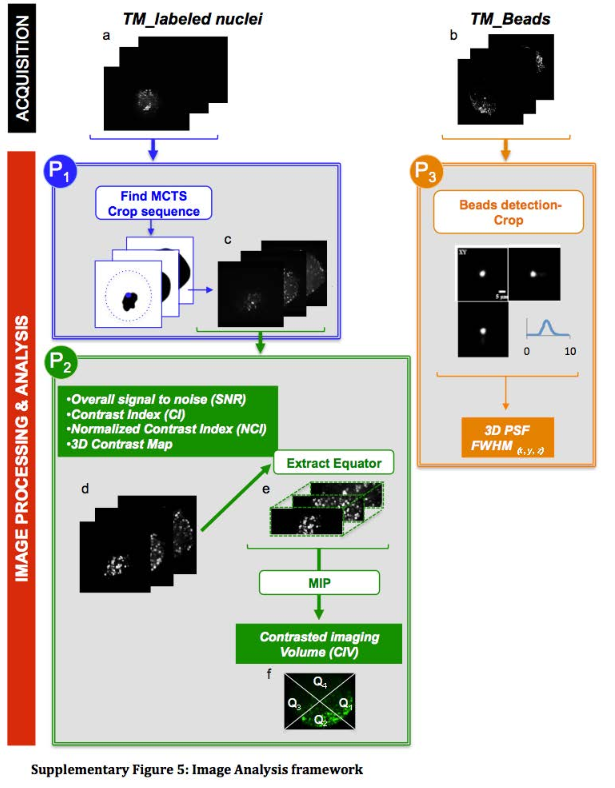
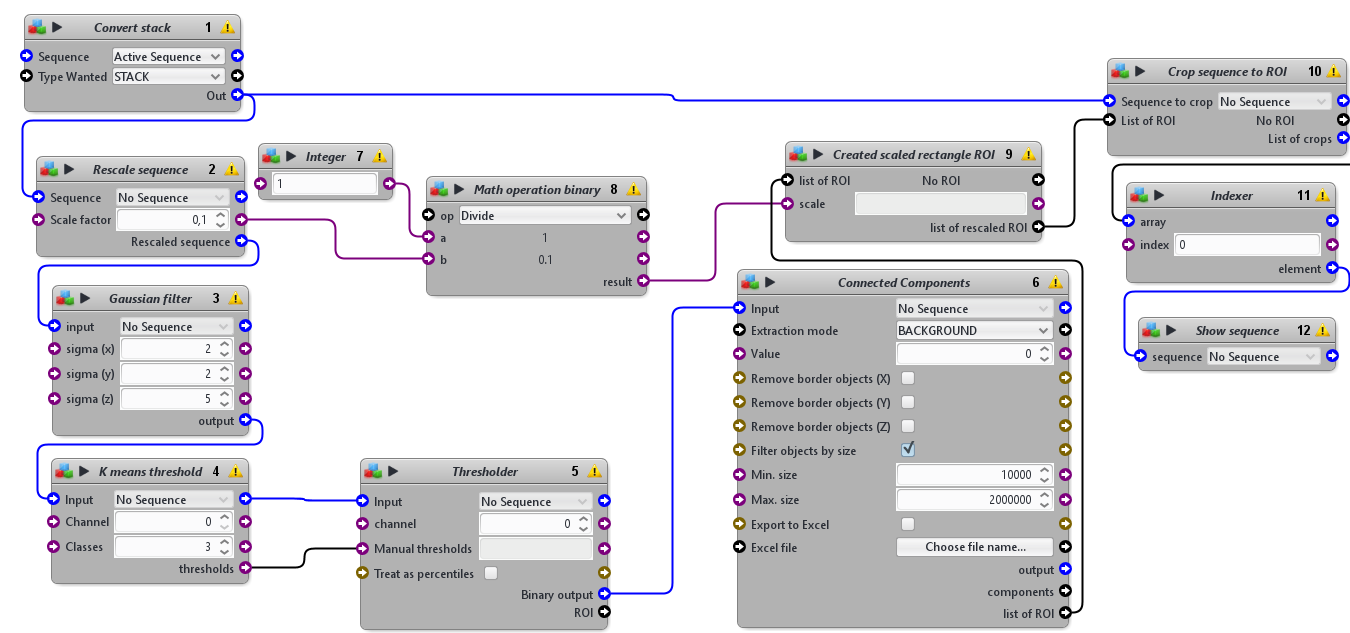
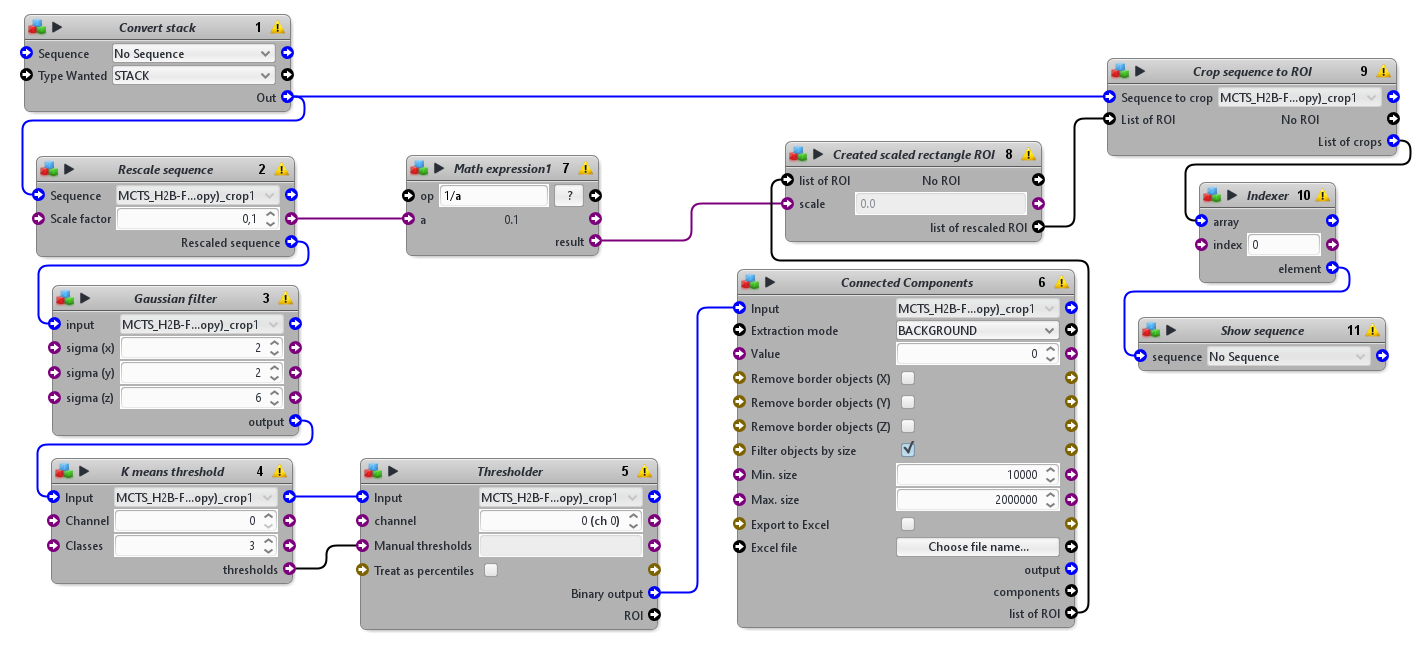
This protocol was designed to to automatically detect the tissue mimics, imaged in 3D with an LSFM modality, and to crop a 3D sub-volume centered on it.
This protocols takes a 3D sequence as input. To detect the tissue mimics, it blurs a rescaled version of the image, apply an automatic threshold to segment the tissue mimics from the background and use connected components to obtain one single ROI delimiting the whole tissue mimics. It then crops the original image using a rescaled version of that ROI.
Please cite Andilla et al. 2017 if you re-use this protocol or make a modified version of it. Figure below comes from the supplementals of Andilla et al., 2017. The protocol described here is the protocol P1.