In this article, we explain how to get help regarding the Icy software, an Icy plugin or protocol, and more generally bioimage analysis with Icy.

Read the documentation
It may seem obvious, but the first thing to do is to look into the documentation.
The Icy download page and the FAQ contain general information regarding installation of Icy, memory usage, publication of protocols or plugins.
Then, there is one documentation page per plugin on the Icy website. Search the plugin name in the search bar of the Icy software, and right click on its name to open the online documentation.
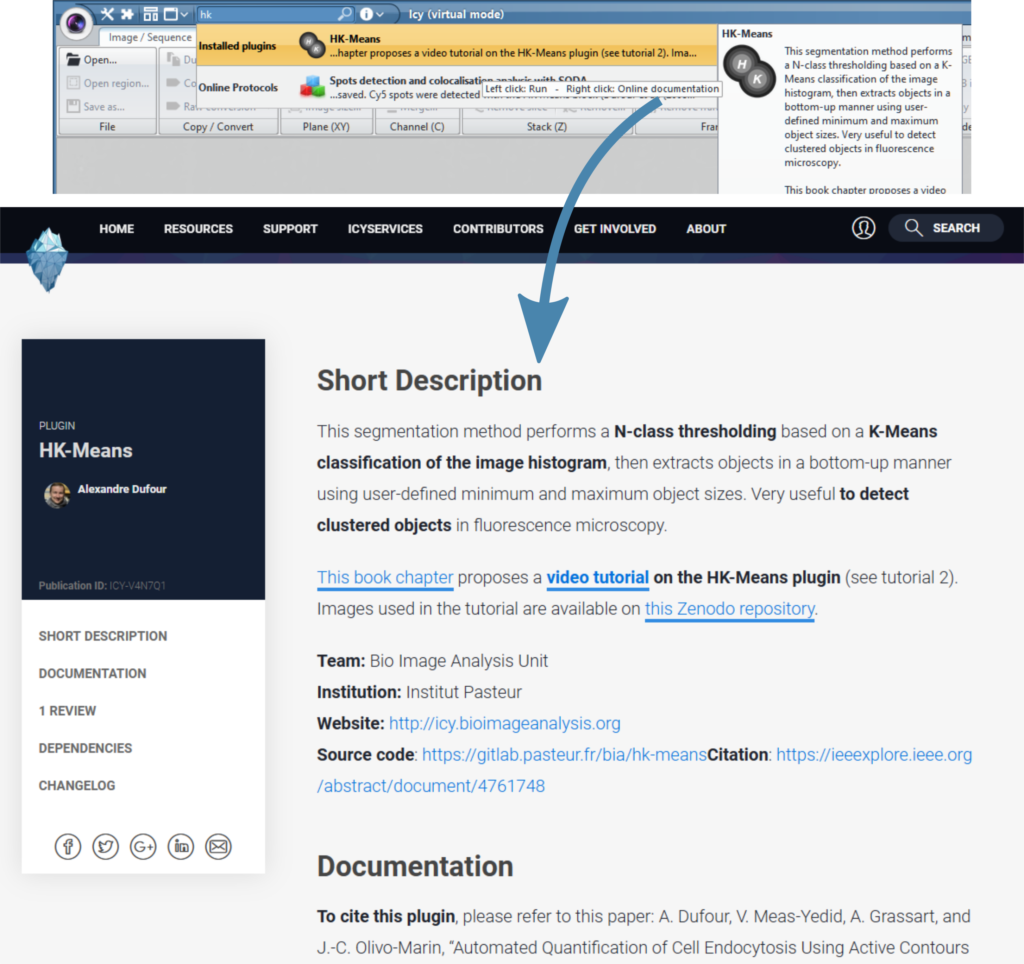
You can also search directly from the website. This can be useful when you are looking for more general information, for instance, you could type “segmentation” to search for plugins which can do segmentation in Icy.
For scripts and protocols creation, we recommend looking at examples of scripts and examples of protocols to get inspired. You can use the search bar on these pages to search only through scripts or protocols.
For plugin developement, you can also search through the Icy javadoc and/or get inspiration by looking at the Icy plugins from the Bioimage Analysis unit that are publicly available on the GitLab server of the Pasteur Institute.
We also gather training materials on the Icy training page. You will find slides, documents and links to online webinars.
Last but not least, all questions asked on the image.sc forum constitute an extra source of information. You can search the forum with keywords and/or with the tag icy.
Troubleshoot the issue
Often, experimenting a bit around a problem can help solve it. As for any kind of scientific experiment, write carefully everything you try. If you ask someone else for help later, this person will be happy to 1. see that you tried by yourself and 2. not have to go through this troubleshooting.
The plugin you use is displaying an error message
Read the error message. Often, the message will be unclear to you because it refers to some Java code of the plugin, but it sometimes contains readable information relative to a specific part of the plugin.
Experiment with the plugin to reproduce the error and answer the following questions:
- What is the sequence of events leading to this error message? In other words, where did you click and in which order? Did you open an image?
- Does it happens all the times or only sometimes? When does it happen, when not?
- Does it happens whatever the type of image (2D, 3D, 3D+time) you use or whatever the type of ROIs?
The protocol you are creating/working with is giving you an error message or producing a different output than what you expect.
If there is an error message, read it. Identify the block(s) that are causing the problem. This block usually displays a white cross over a red disk in its upper right corner as a warning sign that the block could not be executed properly.
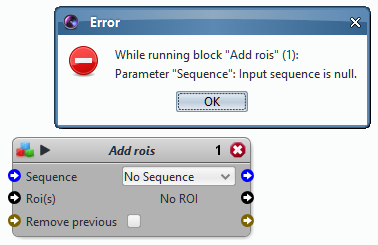
Experiment with the protocol to reproduce the error and answer the questions below.
Note that you surely don’t want to process a folder with 100 images each time you troubleshoot your protocol or go through 7 steps before observing the error. If your protocol is complex, isolate a minimal group of blocks surrounding the blocks that are causing the issue and play only with these blocks. You can save intermediate images and ROIs to use them as input to your minimal workflow.
- Did you reset the blocks before running the protocol? We recommend to always reset, unless some blocks take a very long time to process. This avoids keeping old wrong results.
- Did you select a sequence (an image) as input of your protocol?
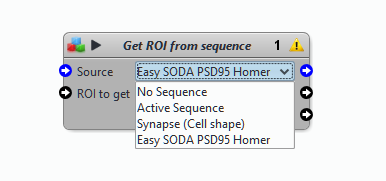
- Did you check all the input and output file paths? If you open a protocol written by someone else, it is likely that the paths are not correct. To check the outputs from a block, use the Display block (right click in the background of the protocol -> Display, or search for the block in the panel on the left of the Protocol Editor)
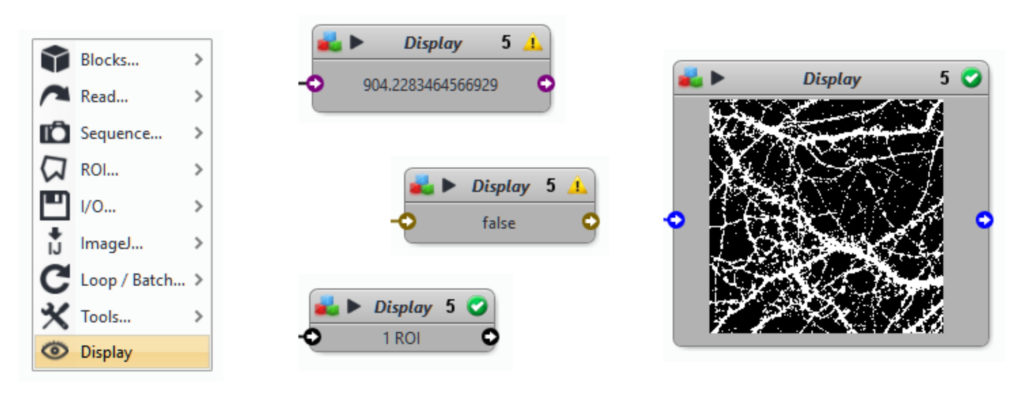
- Are all the blocks connected to the correct inputs and outputs?
- Is the execution order of the blocks ok? Sometimes a block gets by accident the wrong position in the protocol. When you run the protocol, it tries to execute this block first but it fails, because its input variable depends on a block that is not yet executed. The order of execution is given by the number in the upper right corner of the block.
Your script is giving you an error message or producing a different output than what you expect.
Read the error message if there is one. Identify the lines of code causing the issue. As for the protocols, if your code is complex, isolate a minimal group of lines producing the error. Check that all the variables are defined, that names of variables, functions… are consistent over your code, check also the length of objects (are you trying to select something at an index that does not exist?).
Ask your question on the image.sc forum
Despite all your efforts to read the documentation and troubleshoot the problem by yourself, you are still needing help.
We recommend to open a new thread on the image.sc forum with the tag “icy”. Please read the guidelines and the FAQ of the forum before posting and make sure a similar topic is not already opened. On the image.sc forum, you are asking the bioimage analysis community, and in particular the Icy users and developers. Keep in mind that all people answering there are doing it on their free time. You might have to wait a couple of days to get an answer, especially during holiday periods (Christmas, summer break…).
We encourage the discussion to stay public. If you encounter a certain issue, others will encounter it as well. By leaving a trace of your discussions on the forum, you are adding documentation for the community and your future self. Open discussions also give the possibility to anyone to contribute (mention that they encounter the same issue, provide help or share opinions for instance) and strengthen the community of users and developpers.
In your thread, provide as much information as possible to give the possibility to helpers to reproduce your issue. All the information you gathered while reading the documentation and troubleshooting your issue are precious, don’t forget to share them with others. If you have several questions, make separated threads.
Use the following categories for the following topics:
- “Image Analysis” -> general questions on how to analyse an image
- “Usage & Issues” -> issues with a plugin, bug report
- “Development” -> feature request
Here is a check list of information you should provide:
- What is your biological question? Don’t hesitate to share an annotated image pointing at structures of interest for you.
- What is your bioimage analysis strategy? Thresholding, segmentation with HK-Means/watershed/active contours, tracking…?
- An example image, representative of your images dataset (neither the best, nor the worse) in a raw format (no .png, no .jpeg, prefer the format given by your microscope)
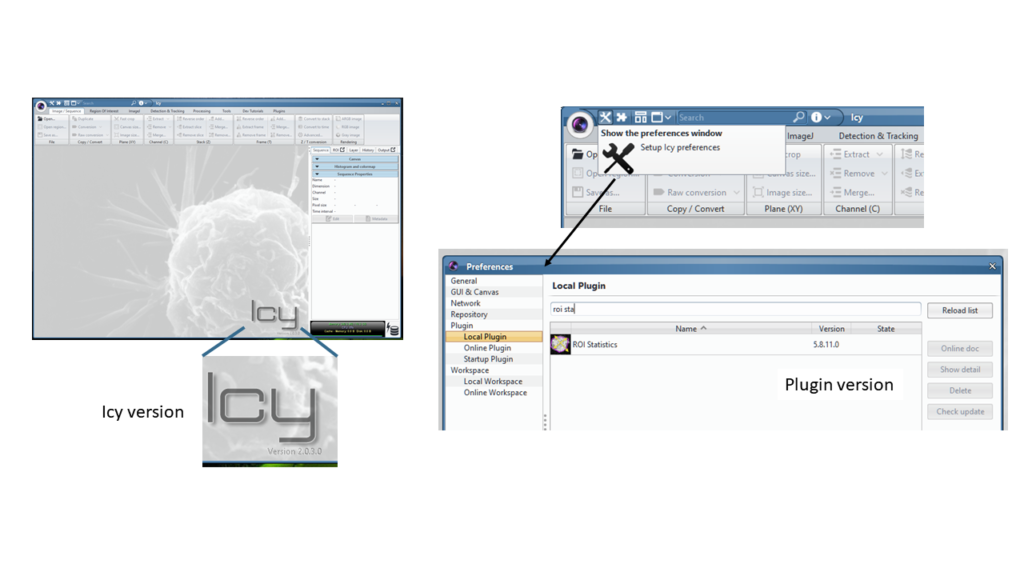
- The tool(s) you used with their version (Icy version 2.1.0.0, plugin HK-Means version…). If you are using a protocol, upload the .protocol file. If you use a plugin, provide the parameters you used to process the example image.
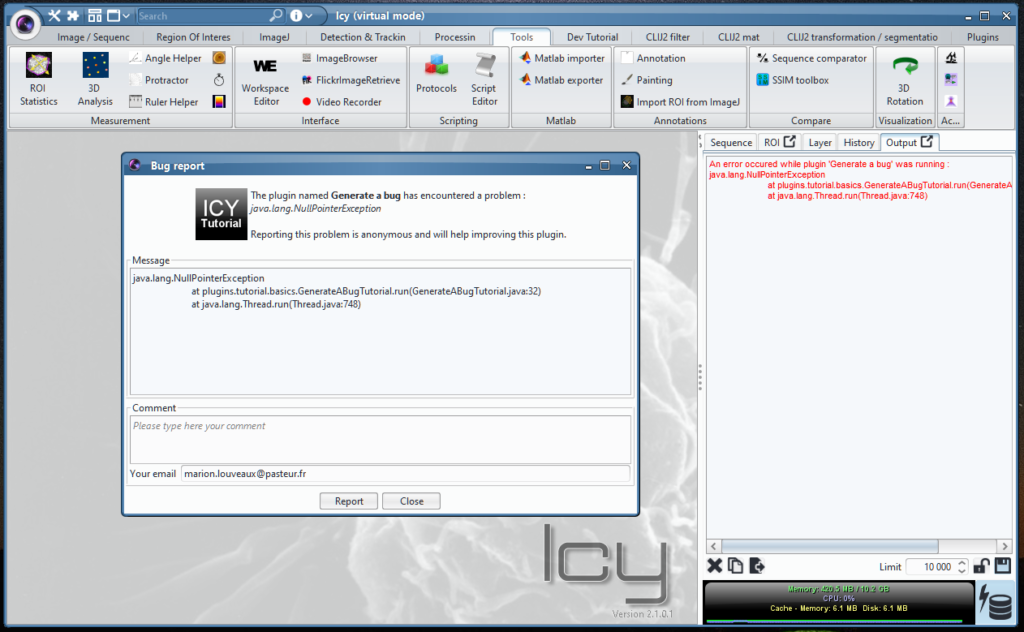
- A copy of the error message or log content (output tab, in the side panel)
If, after around 10 days no one answered your question:
- Add details, try to rephrase your issue
- Contact by email the authors of the plugin or protocol block that is causing your issue and invite them to join you on the image.sc forum (to keep the discussion public). The authors of a plugin are listed on the documentation page of the plugin, click on their name to see their email address. Often, blocks share the same name as their parent plugin. In this case, contact the authors of the parent plugin. If your question is more general, please contact the Icy team by email, indicating the link to the thread you opened on the image.sc forum.