In Icy, “protocols” are bioimage analysis workflows written with a graphical programming language. Here, I detail how to publish and share a protocol on the Icy website.
The procedure in three steps
You can upload a protocol on the Icy website following these three steps:
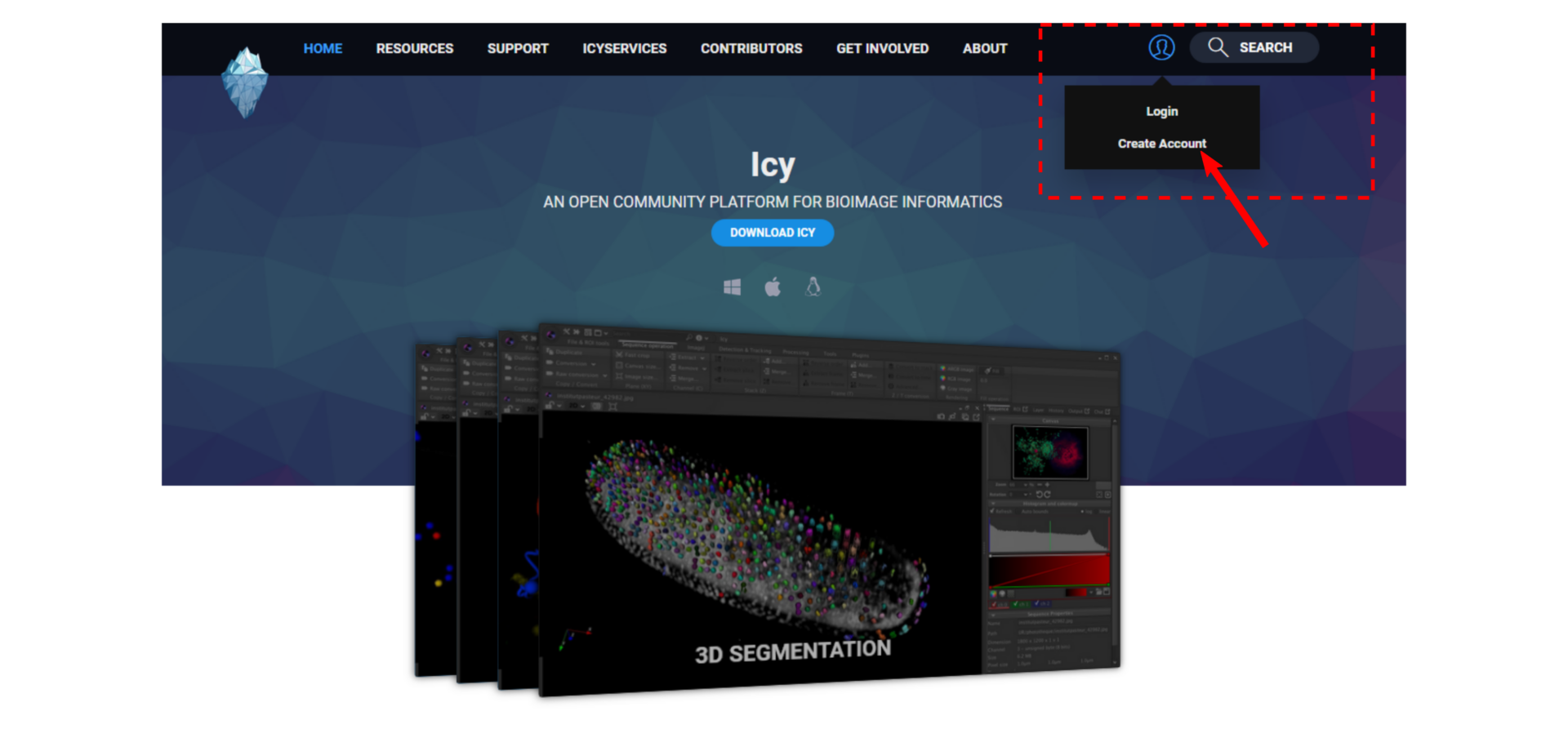
- Create an account for each author of the protocol. Please indicate your real name and prefer one account per physical author rather than one account per lab or team.

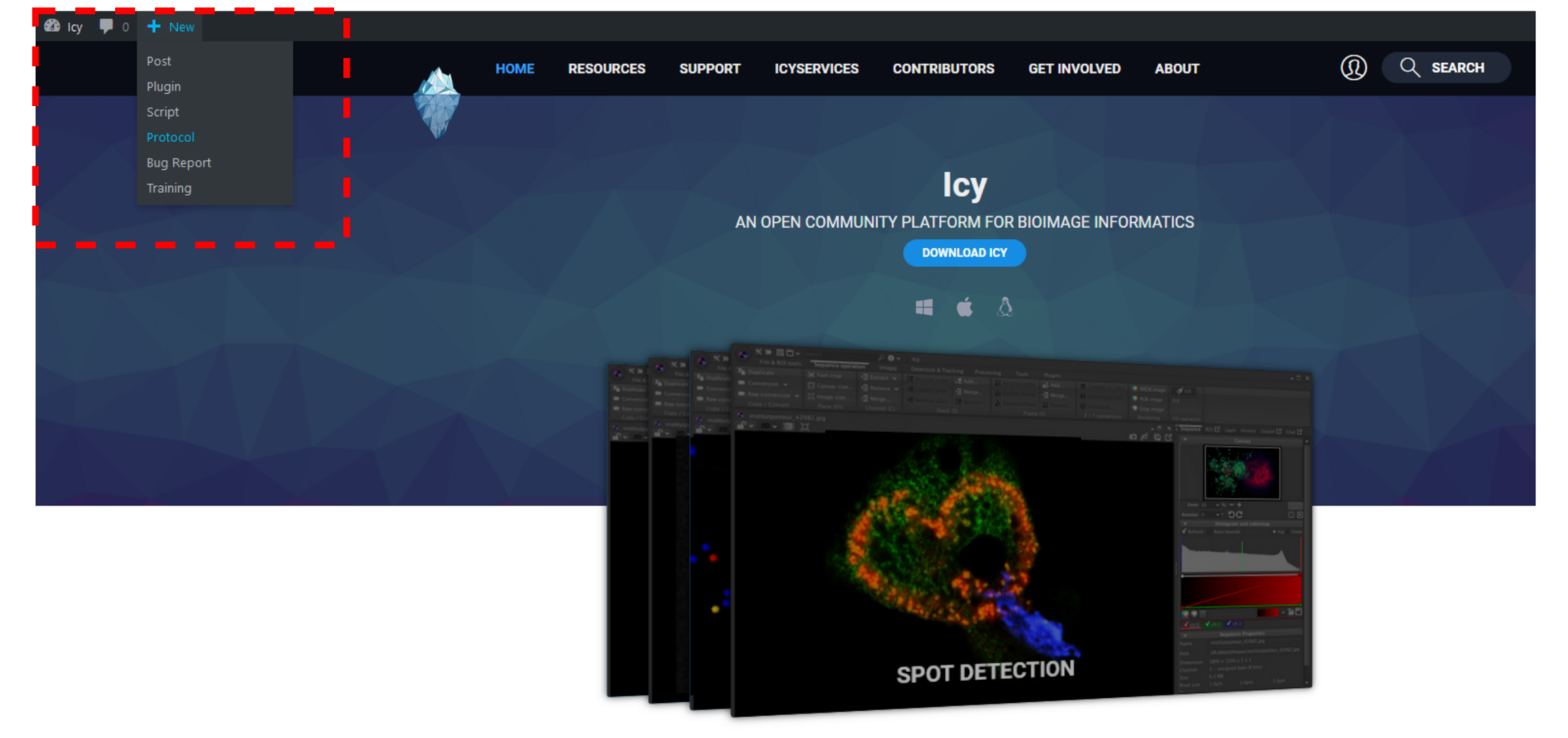
- Log in your account, click on “New -> Protocol“.

- Fill the description and upload your protocol. Below are details on the different sections of the description form.
Why publish a protocol?
Protocols are proper bioimage analysis workflows and deserve visibility. Publishing gives visibility to your protocol in the bioimage analysis community: published protocols can be found on the Icy website via a keyword search and downloaded for reuse from the search bar of the software.
Publishing your protocol contributes to a more transparent and reproducible research.
Do you need some inspiration before getting started? Here and here are two examples of protocols published in the Icy protocol section.
How to fill the protocol description
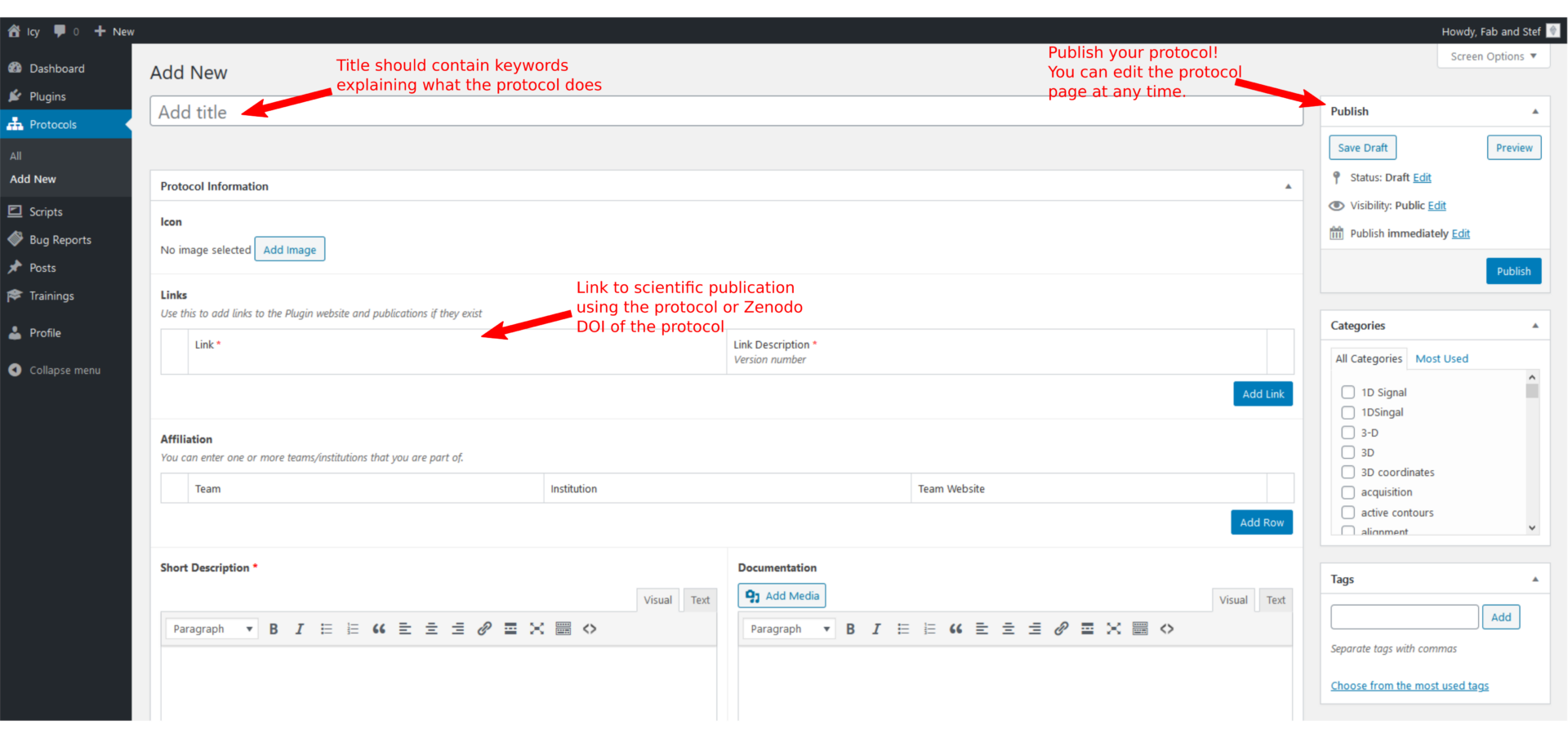
Title: Meaningful but not too long. This title will be used to define the URL of your protocol.
Icon: Usually protocols don’t have an icon, but you can let your creativity speak and add a small meaningful image here to describe your protocol. Otherwise, you’ll get the default protocol icon.
Links: Add links to a reference publication and/or a Zenodo repository hosting your protocol or test images. We encourage you to link your Zenodo upload to the Icy Bioimage Analysis software Zenodo community. Please help other users find easily how to cite your protocol by writing as much information as needed in the description.
Affiliation: The affiliation of all the authors at the time the protocol was written.
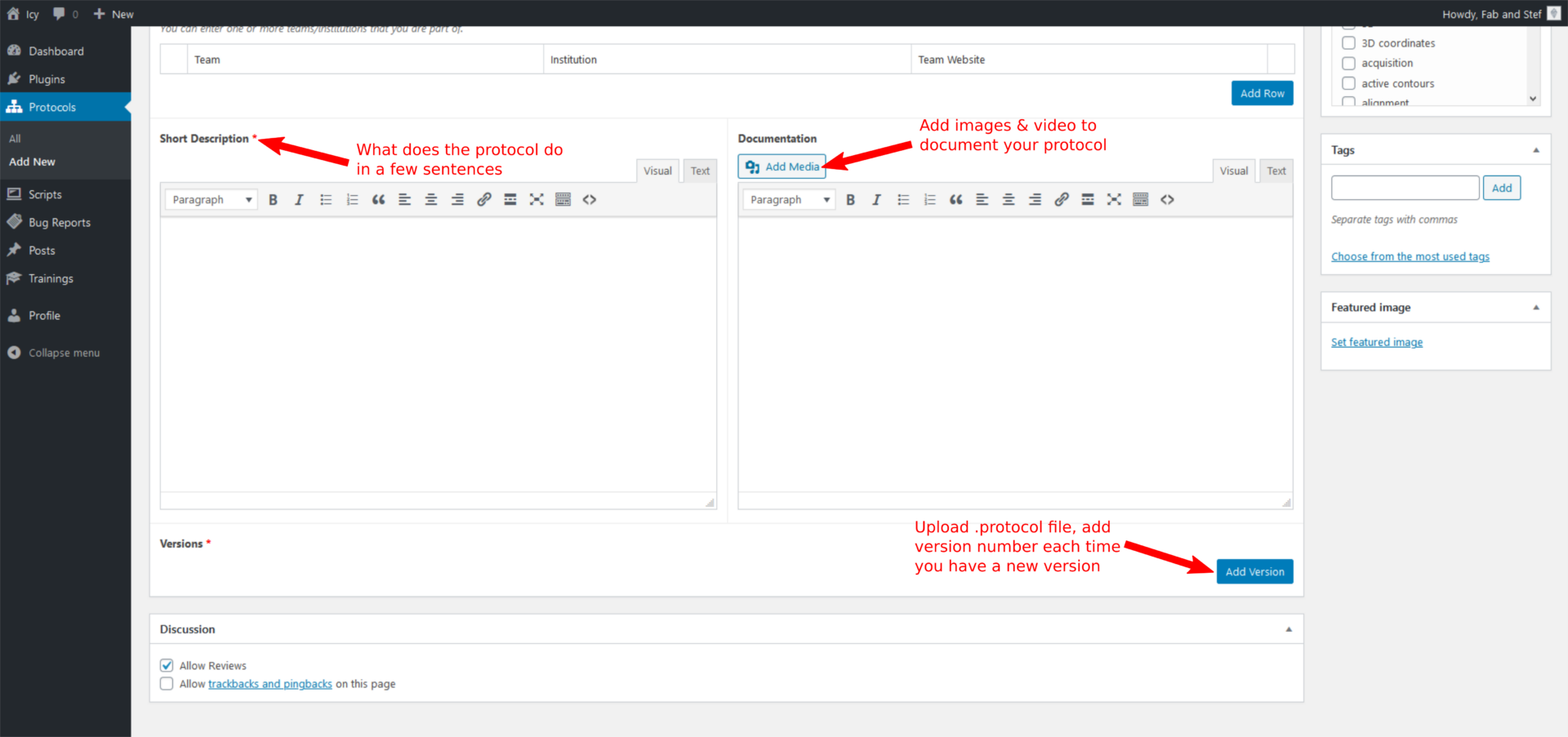
Short description: This helps a lot other users to understand what your protocol is doing. This will also appear under the plugin title in a Google search.
Documentation: Add details on your protocol, such as explanations on the type of image your protocol can process, the custom JavaScript blocks you added, or any other useful tip to run your protocol (size or type or sequence, input variables such as a folder path…). Add screenshots of your protocol or of part of it to describe it more in details than in the version image, as well as bioimage analysis workflow schemes and snapshot of images processed with the protocol. Don’t hesitate to add also video tutorials. I recommend adding link to test images available in public archives, such as Zenodo or the Bioimage Archive.
Versions: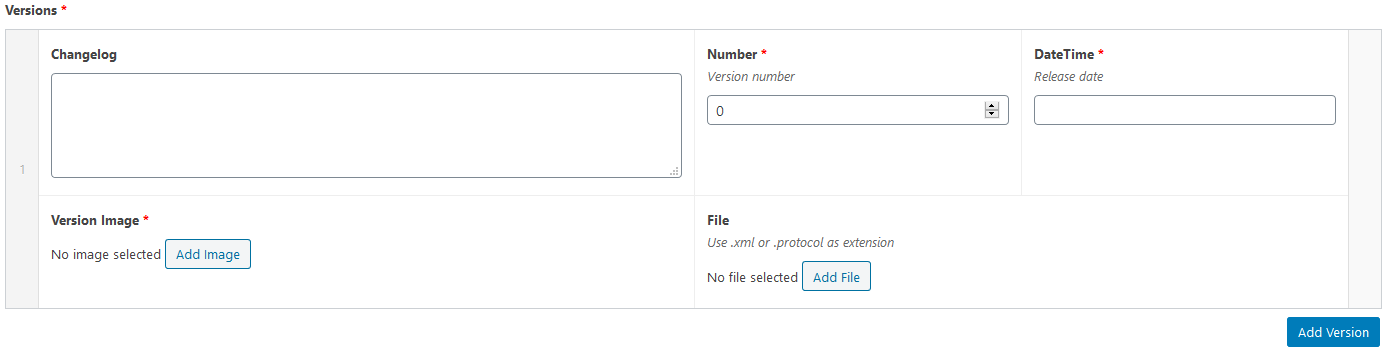
- Click on “Add file” to upload your protocol.
- Add a release date. Unless you have good reasons to do it differently, choose the date of the current day.
- Add a version number. For softwares, versions are usually defined with 3 numbers. The first one indicates the major number version. The second, the minor number version and the third one the patch number version. For instance, version 2.4.3, means that we are using the major release number 1 (major means that a lot of modifications were added compared to the major version 1), minor release number 4 (minor modifications were added compared to version 2.3) and patch number (third patch done on the version 2.4, a patch is a small modification correcting one given bug). More details on software versionning here. For Icy protocols, use 1.0 for the first version you release, and then 1.1 if you release a second version (= you upload a new version of your protocol on this page) containing minor modifications such as a change of parameter value, and 2.0 if you add more consequent modifications such as adding a block or several blocks.
- Changelog: Write “first release” if this is the first time you upload this protocol. Otherwise, give details on what has changed compared to the previous version.
- Version image: Snapshot of the protocol.
- Click on Add version to add a new version row.
Categories
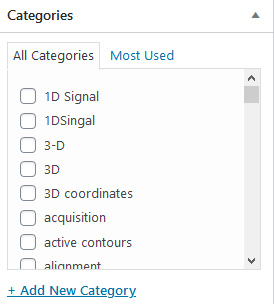
Choose the most meaningful keywords to describe your plugin. These categories are used by search engines to find your plugin.
Tags
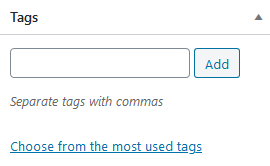
Similar to categories, leave empty or choose the most meaningful keywords to describe your plugin. These tags are used by search engines to find your plugin.
Discussion
Accept or not reviews from users.
Publish
Click on publish when you are ready to release your protocol. You can save the draft for later and preview the changes. Note that you can modify this page at any time after publication.
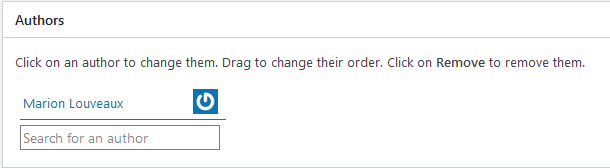
How to add or modify authors
After publishing the protocol, you can still add authors to the protocol. All authors need to have a user account on the Icy website.