Short Description
This protocol allows you to segment Cell shape thanks to the HK-Means method. It will create an overlay of the segmented results over your original picture as a ROI.
Citation (Zenodo): https://doi.org/10.5281/zenodo.4317782Documentation
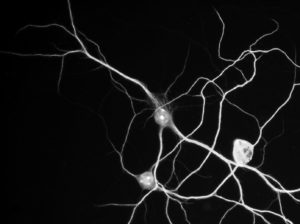
Let’s say that you need to segment this cell.
You just have to :
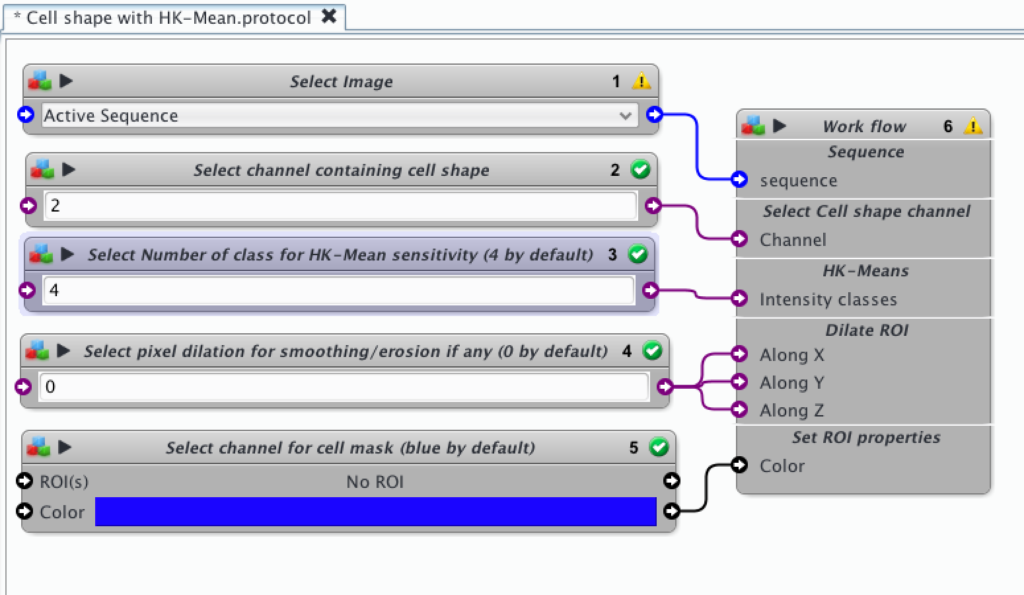
- select the desired file in block 1.
- Select the channel of interest for cell labeling (here channel 2)
- Select the number of classes you want to use in HK-Means segmentation. 2 classes would correspond to conventional automatic thresholding. By increasing the number of classes you increase the sensitivity and go deeper in the faint regions. By default this value is set up at 4.
- You can add a facultative step of dilation/erosion that will smooth the mask. By default it is set to 2 pixels.
- Choose the color for your ROI. By default it is set to blue, but you can choose your favorite one.
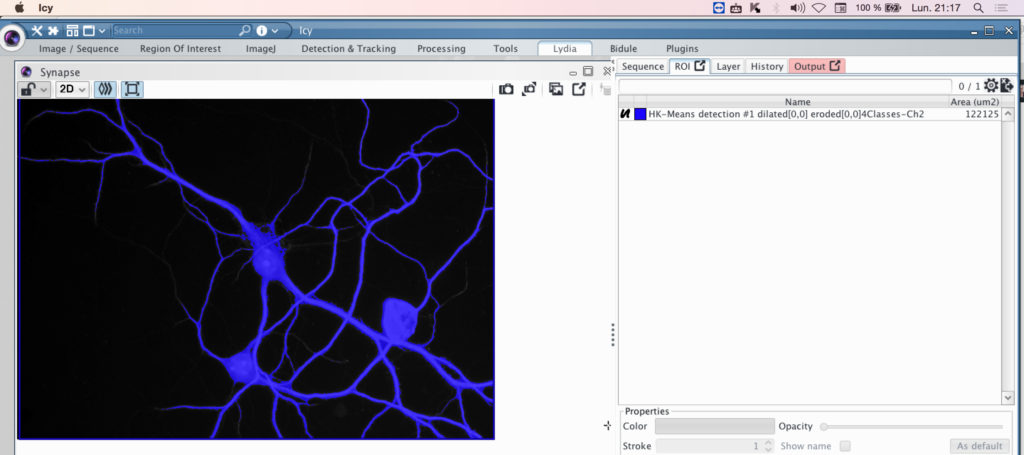
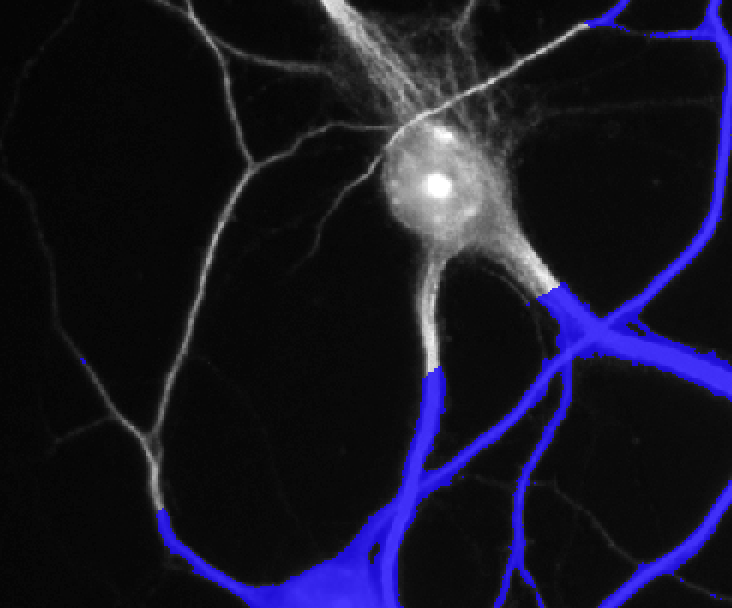
Here is the Cell shape ROI in blue overlaid on the original picture. You can mask the ROI by clicking on the button indicated in red below.
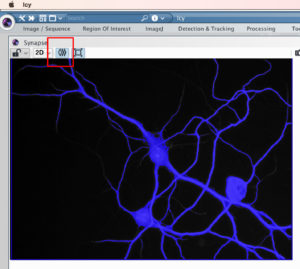
The name of the ROI with the parameter you have selected will appear in the ROI panel.
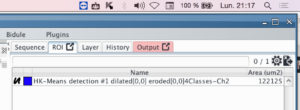
Since the ROI’s name depends on the parameters, you can run the protocole several time without crushing your different ROI.